SNF3 and YDL199c
This web page was produced as an assignment for an undergraduate course at Davidson College
As discussed in previous web assignments (click here to access), SNF3 is found on the fourth chromosome of Saccharomyces Cerevisiae. It is located in the plasma membrane and encodes for a glucose sensor protein. It is involved in glucose binding and glucose transporter and receptor activity. This web assignment will examine the available proteomic databases with the search term of SNF3 to determine the protein function, location, and structure.

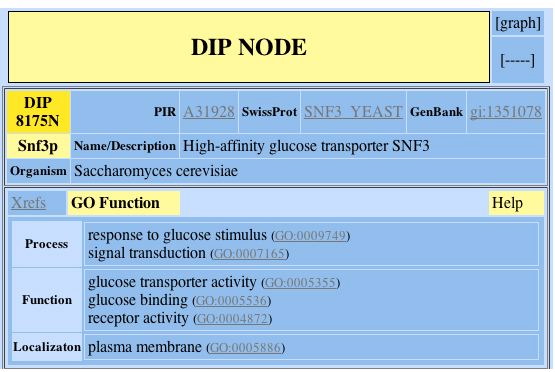
Figure 1. DIP NODE database's GO function. This figure reveals the process, function, and localization of the SNF3 protein in the Saccharomyces cerevisiae (DIP 2004). Image located at <http://dip.doe-mbi.ucla.edu/dip/DIPview.cgi?ID=DIP:8175N&MD=2>. (permission pending)
Searching the DIP database produced no graph of the SNF3 protein interactions, although it did give the function, process, and localization of the protein. As discussed in previous web assignments and confirmed in this figure, the protein for SNF3 is located in the plasma membrane and acts as a glucose receptor in response to glucose stimulus(DIP 2004).

Figure 2. MIPS database lists the functions of the SNF3 protein. This figure shows the functions that have been experimentally determined for the SNF3 protein and suggests some protein interactions (MIPS 2004). Image located at <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YDL194w&db=CYGD>. (Permission pending)
| This figure reveals some additional functions this protein may have, such as involvement in the biogenesis of the cell wall and expression of the C-terminal domain. The FunCat section reveals the involvement of the protein in metabolism, cellular transport, and signal transduction. It is also found that there are functions that directly affect other proteins and thus suggest an interaction with the proteins of HXT5, HXT6, and HXT7, SUC2, and HXT2. For some of these interactions, SNF3 is required for induction of another protein, and at other times it is needed for the reduction of protein. This leads me to believe that in different environments and in the presence of different proteins, SNF3 has different effects. This database also provides the isoelectric point of SNF3, which is 5.22 (MIPS 2004). The isoelectric point is equal to the pH of the environment when the protein is neutral (Campbell 2003). This means that the protein is neutral in a slightly acidic environment. |

Figure 3. MIPS protein-protein interactions. This figure shows the protein-protein interactions of SNF3 and other proteins (MIPS 2004). Image found at <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YDL194w&db=CYGD>.
| This shows that SNF3 has a physical interaction with the protein MTH1 and genetic interactions with CYC8, HXT1, HXT2, HXT3, HXT4, MSS2, RGT1, RGT2, and SKS1. Although this database is the same one used to generate Figure 2, the results are different. The only protein-protein interaction that is listed in both parts is the interaction of SNF3 with HXT2. The interactions from Figure 2 may not be direct interactions, but merely associations between the protein (MIPS 2004). |

Figure 4. MIPS localization of the SNF3 protein. This figure shows that the SNF3 protein is located in the plasma membrane, cytoplasm, and the endoplasmic reticulum (MIPS 2004). Image found at <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YDL194w&db=CYGD>.
| This figure confirms that the SNF3 protein is found in the plasma membrane, but it also reveals that the protein is also found in the cytoplasm and the ER of the cell. This helps to explain the interaction of SNF3 with the protein SKS1, which is found in the cytoplasm. |
The SGD database did not provide any new, relevant information about the protein. However, it contained a section that involved searches of other databases for the protein interactions. There were four different searches, one of which was the DIP search as done above. The search of the Portal Pathcalling database did not return any SNF3 interactions. Here are the images of the other two database findings.
Here are the 11 protein-protein interactions found by the database BIND. More information about their functions, processes, and locations can be found in figure 6.
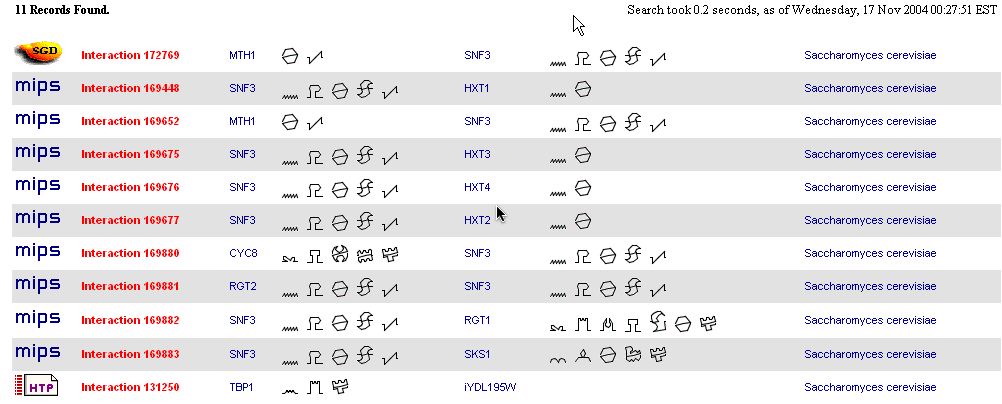
Figure 5. The BIND results of SNF3 protein-protein interactions. This figure shows 11 found protein interactions of SNF3 (YDL194W) with ten different proteins (BIND 2004). Image found at <http://bind.ca/Action?textquery=RecordType:%20(interaction%20complex%20pathway%20)+AND+YDL194W>. (Permission pending)
This database includes interactions with nine of the same proteins found in the MIPS database. However, it is interesting to note that the protein MTH1 interacts twice with SNF3. I doubt that this is a typo, but there may be a few ways to explain it. One way is if there were two different experiments that found this interaction and this database includes them separately. Another possibility is that the proteins interact in two different locations and thus have two totally different interactions. |
At first, it was unclear why the interaction between TBP1 and iYDL195W was listed in the same results as the reactions of SNF3, or YDL194W. However after looking at the link for iYDL195W, it seems as though this segment of DNA regulates SNF3 (see below figure).
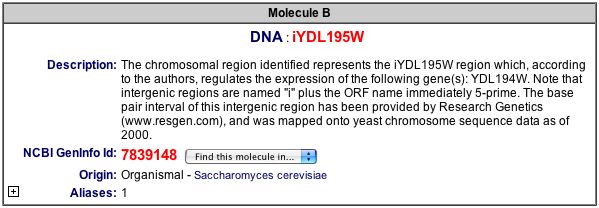
Figure 6. Description of the intergenic DNA region, iYDL195W.This figure gives a brief explanation of how the iYDL195W is related to SNF3, or YDL194W (BIND 2004). Image found at <http://bind.ca/Action?pg=3001&identifier=bindid&idsearch=131250>. (Permission pending)
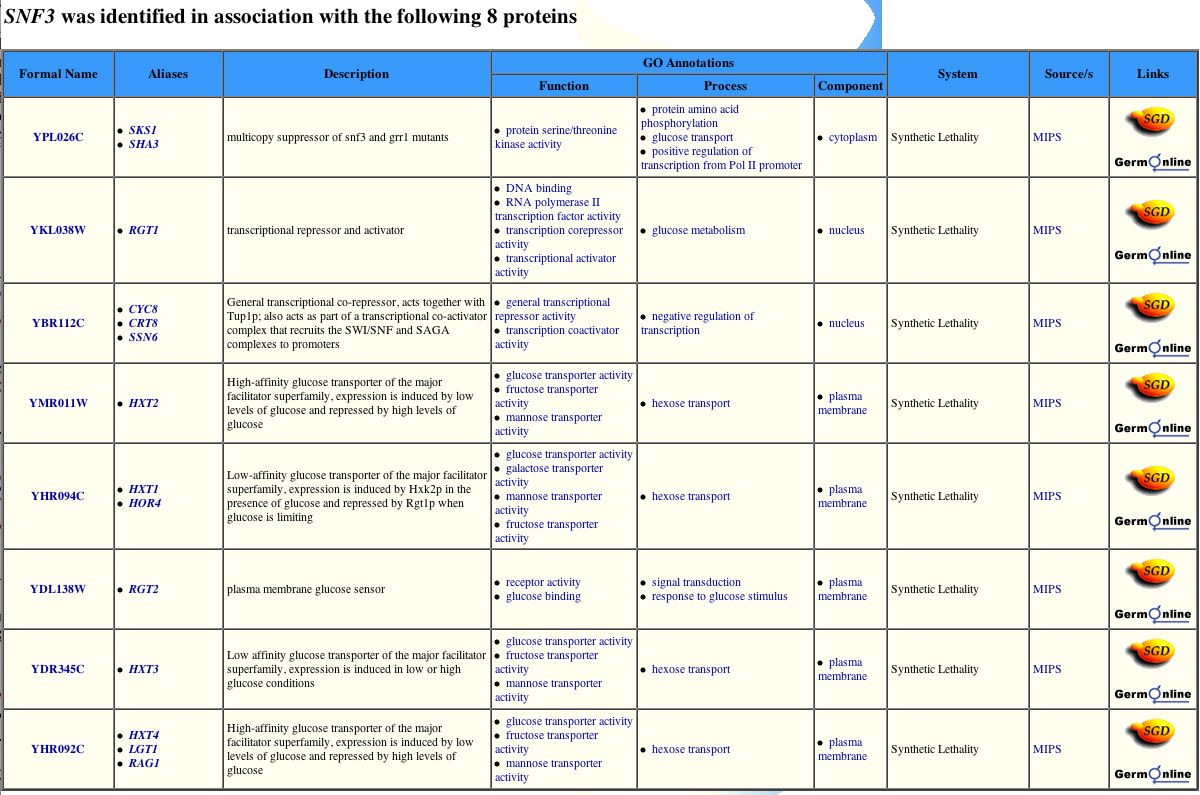
Figure 7. The GRID results of SNF3 protein interactions. This image provides eight proteins identified with SNF3 and information regarding those proteins (GRID 2004). Image found at <http://biodata.mshri.on.ca/yeast_grid/servlet/SearchResults?keywords=YDL194W>. (Permission pending)
Although this database shows that SNF3 interacts with eight of the same proteins, it fails to recognize MTH1 as a protein that interacts with SNF3 (GRID 2004).
| This is interesting, especially because this protein was the first protein, and only protein listed, in the MIPS database and listed for two different interactions with SNF3 in the BIND database. This may be because the GRID database used different experimental conditions to search for the protein-protein interactions. Seven of these eight proteins are described as dealing with similar functions as the SNF3 protein and are located in either the plasma membrane with the SNF3 protein or in the nucleus. However, the protein SKS1 does is involved in the process of amino acid phosphorylation as well as glucose transport. This suggests that SNF3 has a slight impact on this process as well as the metabolism and transportation of sugars. |
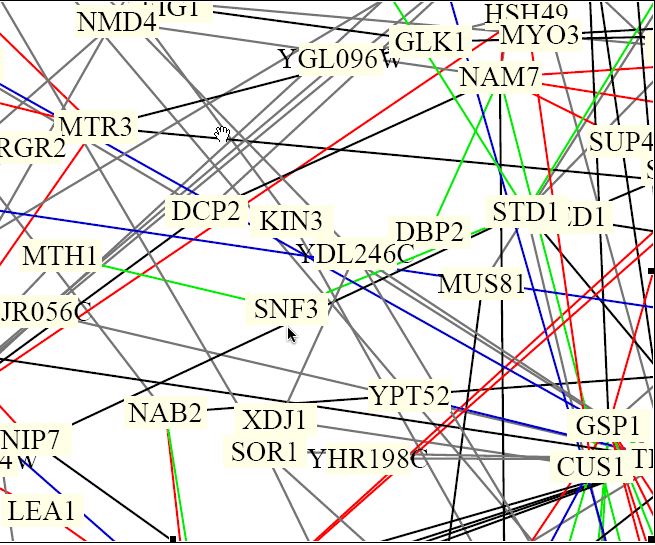
Figure 8. Benno Schwikowski protein interactions of the yeast genome. (Schwikowski, et al. 2000) This figure is only a section of the full diagram of 2,358 interactions between 1,548 proteins. Full image found at <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/seq/Benno/NB_Figure1color.pdf>.
The protein SNF3, which is located near the center by the pointer arrow, seems to have two green lines that connect to it. The green lines are coded to mean that the interactions have cellular roles that are the same the their localizations differ. The interactions for SNF3 seem to be to the protein MTH1, which has been noted by the MIPS and BIND databases as well, and to STD1 protein (Fields, Uetz, and Schwikowski). The protein STD1is a "dosage-dependent modulator of glucose repression" (MIPS 2004).
| The interaction between SNF3 and STD1 is not necessarily a true interaction. The way that this figure is set up reveals that too much information is not necessarily a good thing. It is difficult to tell if the SNF3 interacts with STD1 or with a protein whose name is underneath that of STD1. The chances, however, that these two proteins do interact is probably high. Since STD1 is involved glucose repression at certain dosages of glucose, and SNF3 is a glucose receptor that is sensitive to low concentrations of glucose, there may be some similarities in their functions. This makes it seem as though it were possible for these two proteins to interact, even though they are not listed as interacting in any of the databases used so far in this analysis. I also found that the expression of SNF3 is likely to be regulated by the intergenic region of DNA iYDL195W. This section of DNA interacts with the protein TBP1, which suggests that there may be some sort of interaction between SNF3 and this TBP1. |
The Aging, Degradation, and Membrane files with links located at http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/chapter6/deluxe.html included the protein SNF3. However, the data was so clumped and crowded that it was impossible to see either the interactions or the protein itself.
These other databases were searched, and either no information was displayed or no new and relevant information was given.
Enzymes and Metabolic Pathways
| Overall, this new data from other databases supports the claims of my earlier web assignments and also suggests a few more ideas. The location is still known to be in the plasma membrane, but now I can conclude that it is also found in the ER and cytoplasm. The cytoplasmic interaction between SKS1 and SNF3 suggests that SNF3 may be involved in the phosphorylation of amino acids and the regulation of the transcription from a promoter. These newly found functions are only an addition to the glucose transporter and receptor functions that were found in previous web assignments and supported by this analysis. |
As hypothesized in previous web assignments (click here to access), the non-annotated gene YDL199C is probably associated with the signal transduction of glucose and possibly associated with the metabolic shift from fermentation to respiration. Just as SNF3, the gene is located on the fourth chromosome of Saccharomyces cerevisiae and the protein is found in the plasma membrane. The search term YDL199C was used as the search term in these databases to examine the protein function, location, and structure of this gene.

Figure 9. MIPS database lists the functions of the YDL199C protein. This figure shows the function that has been determined for the YDL199C protein (MIPS 2004). Image found at <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YDL199c&db=CYGD>.
This figure explains that the protein of YDL199C functions in cellular transport of carbohydrates and is in the sugar transporter family. This data is coincides fairly well with the data found in the previous web assignments. These assignments concluded that the protein was involved in the signal transduction of sugars, which may be related to the transport routes evident from this database.

Figure 9. The MIPS localization of the YDL199C protein. This figure shows that the protein of YDL199C is located in and integral membrane and the mitochondria (MIPS 20004). Image found at <http://mips.gsf.de/genre/proj/yeast/searchEntryAction.do?text=YDL199c&db=CYGD>.
This figure is interesting because previous searches have not given the localization of this protein in the mitochondria, only in the plasma membrane. And although this figure shows that the protein may be in the plasma membrane, it is "not assigned to a specific membrane" and may be in any integral membrane as well. The database does not provide any protein-protein interactions for this protein. (MIPS 2004).
| It is easy to believe that with these newly found possible locations of the protein, there may be many interactions. The more places that this protein resides, the more chances it has to interact with other proteins. However, if this is true, then these interactions have yet to be found. It is also interesting that this protein is found in the mitochondria because the mitochondria is a structure that helps break down glucose in order to get more energy for the cell. When the protein is located inside the mitochondria, it may have a similar function of sugar transport that it does have outside of the mitochondria. |
As was the cases with SNF3, the SGD database did not provide any new, relevant information on the YDL199C protein. It also contained links to searches of protein-protein interactions in other databases. There were four other databases, three of which did not provide any interactions of YDL199C. The other database finding is below.

Figure 10. The BIND results of YDL199C protein-protein interactions. This figure shows one interaction between the intergenic region of DNA that controls the expression of YDL199C and the protein TBP1 (BIND 2004). Image found at <http://bind.ca/Action?textquery=RecordType:%20(interaction%20complex%20pathway%20)+AND+YDL199C>.
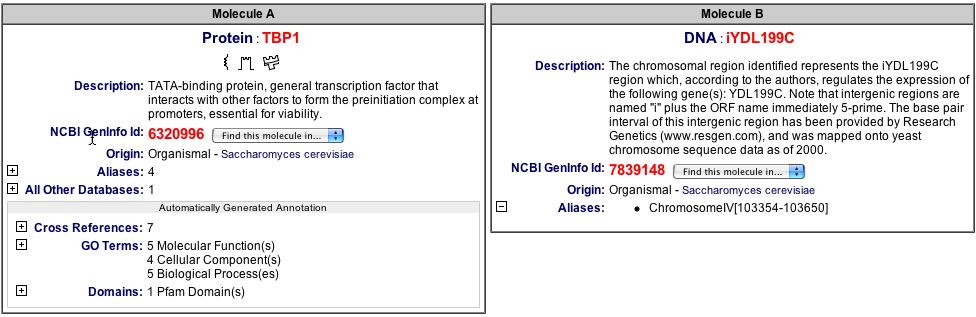
Figure 11. Explanation of the TBP1 and iYDL199C protein-protein interaction. This figure describes each of the components in this interaction (BIND 2004). Image found at <http://bind.ca/Action?pg=3001&identifier=bindid&idsearch=131252>.
| It is worthy to note that there is only one interaction and it does not involve the actual protein, but it's regulator. This is hard to believe, considering the location of the protein is in a membrane or the mitochondria. These locations tend to be part of complex processes that are involved with many proteins, yet there are no known protein-protein interactions for this specific protein. This may be because of a lack of research on this particular protein to this point or there really may not be any interactions. |
These other databases were searched, and either no information was displayed or no new, relevant information was given.
Enzymes and Metabolic Pathways
After searching these proteomic databases, the hypothesized function of the protein remains about the same. Earlier searches led to the conclusion that it was involved in the signal transduction of glucose, and these searches confirm the involvement with glucose. There were, however, some differences in the cellular location. In the earlier analysis, it was thought that this protein was located only in the plasma membrane, but these searches have led to the finding that the YDL199C protein is also located in the mitochondria and other integral membranes; however this protein was not located in the pdf file of membrane protein-protein interactions. The lack of protein-protein interactions of this protein was very surprising. Whether it be because of lack of research to this point or merely the lack of interactions of the YDL199C protein, this result was not expected. With these differences in data and lack of data, it is only fitting to continue the research and experiments with the protein to learn more about it. The following are just some of the many possible experiments that can be run to increase the knowledge about the protein YDL199C. One of the problems that arose during these searches was the different localizations of thee protein within the cell. To test whether all of these locations valid or not, an immunoflourescent antibody able to bind to the protein YDL199C can be used. Once it binds to the protein, you can look at the localization of the tagged proteins. In this case, I would expect a fair amount of the antibodies to be in the plasma membrane, and some in the mitochondria and other membranes as well. Wherever there may be glucose transport will probably attract the antibodies. Another experiment to run is the yeast two-hybrid method to test for protein-protein interactions. The YDL199C protein would be the bait protein bound to the DNA Binding Domain. A variety of other different proteins can act as the prey protein bound to the Activation domain. When they are separate, neither complex can initiate transcription; however, when they are produced in the same cell, they will interact and transcribe a reporter gene. I would expect proteins that are involved in the production, transportation, or metabolism of glucose to be able to bind with the YDL199C complex to initiate transcription. Also, the proteins involved in fermentation, respiration, or the shift from one to the other might be able to combine to initiate transcription. It would also been beneficial to know how much of this protein would be produced in different environmental conditions, specifically at different glucose concentrations. Chait's method using stable isotopes would be beneficial for this experiment. I would grow three different samples of cells in three different populations of different nitrogen isotopes. One sample would be the low concentrations of glucose, one could be average concentration, and one could be high concentration. I would expect this experiment to show less protein transcribed in the low concentrations of glucose because there would be less glucose to signal for. I would expect the amount of the YDL199C protein to correspond with the amount of glucose present in the environment. It would also be interesting to know if this protein was vital to the life of the cell. To do this, you would create a knockout group of cells that contained none of the YDL199C protein and grow them under different conditions. If the cells all die, then this protein must be vital. However if nothing happens, there may be other proteins that are able to pick up the slack of the missing YDL199C protein. If this happens, it would be important to see which protein did extra work. |
[BIND] Biomolecular Interaction Network Database. 2004. <http://bind.ca/index.jsp?pg=0>. Accessed 17 Nov 2004.
Campbell, Malcolm A., and Laurie J. Heyer. Discovering Genomics, Proteomics, and Bioinformatics. CSHL Press: San Francisco, 2003.
[DIP] Database of Interacting Proteins. 2004. <http://www.yeastgenome.org/>. Accessed 17 Nov 2004.
Dolinski, K. et al. (2003). Saccharomyces Genome Database. <http://www.yeastgenome.org/> Accessed 18 Nov 2004.
ExPASy Proteomics Server. 2004. <http://www.expasy.org/>. Accessed 17 Nov 2004.
[MIPS] Munich Information Center for Protein Sequences. 2004. <http://mips.gsf.de/genre/proj/yeast/index.jsp>. Accessed 18 Nov 2004.
[PDB] Protein Data Bank. 2004. <http://www.rcsb.org/pdb/index.html>. Accessed 18 Nov 2004.
Portal Pathcalling. 2004. <http://portal.curagen.com/pathcalling_portal/index.htm>. Accessed 17 Nov 2004.
Schwikowski, B., Uetz, P., Fields, S. 2000. A network of protein-protein interactions in yeast. Nature Biotechnology.18: 1257-1261. <http://occawlonline.pearsoned.com/bookbind/pubbooks/bc_mcampbell_genomics_1/medialib/seq/Benno/NB_Figure1color.pdf>. Accessed 17 Nov 2004.
Swiss-2D Page Map Selection. 2004. <http://ca.expasy.org/cgi-bin/map1>. Accessed 17 Nov 2004.
TRIPLES Database. 2004. <http://portal.curagen.com/pathcalling_portal/index.htm>. Accessed 17 Nov 2004.
Yeast GRID. 2004. <http://biodata.mshri.on.ca:80/yeast_grid/servlet/SearchPage>. Accessed 17 Nov 2004.
Davidson College Biology Homepage
Davidson College Genomics Homepage
Email any questions or comments
© Copyright 2004 Department of Biology, Davidson College, Davidson, NC 28036
Send comments, questions, and suggestions to:macampbell@davidson.edu #